Electrical engineering sparks discovery in how human nuclear receptors bind to DNA
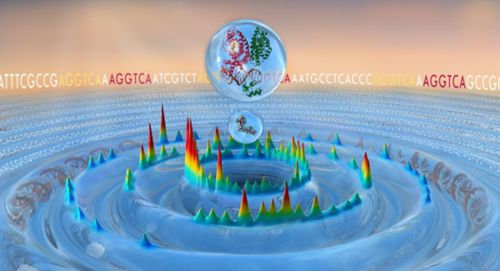
Hidden nuclear receptor binding sites reside in the genomic ocean of DNA sequences. Figure by Madison Rice, St. Jude Department of Chemical Biology & Therapeutics
A traffic light sends us signals that we interpret to know when to advance, slow or stop our car. Similarly, our body requires signals to tell it how to function. Binding, the literal process of one substance attaching to another, often serves as this signal. Proteins bind to DNA, RNA, metabolites, or other proteins to drive distinct functions, sometimes directly but also as the first domino in a long pathway, triggered by a cascade of binding events.
Such is the case for nuclear receptors, a class of proteins that are responsible for sensing signaling molecules, such as hormones, metabolites and drugs to elicit a physiologic response. This family of 48 proteins and their myriad “isoforms,” different types of the same protein, are composed of two distinct binding modules: a ligand binding domain at one end and a DNA binding domain at the other.
When hormones such as estrogen bind the ligand binding domain of the estrogen receptor, this acts as a signal for the receptor to move inside the nucleus of the cell. At this stage, the DNA binding site becomes key, because it is naturally designed to bind to a particular set of sequences of DNA. These sequences serve as controllers for regulatory elements of genes involved in a range of functions, including cell division, which when unchecked leads to several cancers. Binding is the signal for the expression of targeted genes, which is the final result in a long journey kickstarted by the first binding event between nuclear receptor and hormone.
Ligand binding is well studied. The interactions of proteins with small molecules or other proteins follow a clearly defined set of rules which can be exploited to design therapeutics. Nuclear receptors are vital in this system, accounting for around one in six of all FDA-approved drugs. Most of these are designed to hit the ligand binding domain. But what is going on at the other end of the receptor, at the DNA binding domain, is more of a puzzle. One that Aseem Ansari, St. Jude Department of Chemical Biology & Therapeutics chair is keen to solve.
“We have been looking at that problem for a while, because nuclear receptors are drivers of a variety of cancers, especially hormone-driven cancers,” Ansari said. “One thing we noticed is the molecules that are designed to try and displace nuclear receptors from DNA are often unsuccessful.”
Current understanding of DNA binding is a drop in the ocean
While there are some noticeable trends, not much is understood about the range of genomic sites where nuclear receptors bind. Confounding this is the fact that despite containing the same genome, a given nuclear receptor in different cells can bind at different genomic locations. “Depending on the cell type, estrogen receptors are often found in different places on the genome,” Ansari explains.
“Same genome, same binding sites. So, why are these receptors binding in different places?” he questions.
Published recently in Nature Communications, Ansari and his team tackled this dilemma. The researchers set out to explore underlying DNA sequence patterns that allow for nuclear receptor binding. When they initially looked in genome databases to capture all the sequences that are involved in binding, they realized the scope of the challenge. “We had a million sequences out of a billion that came back,” Ansari said. “You can start to assign these by seeing some relationships amongst sequences that are most frequently associated with the receptor, but by the time you get to the very bottom of the enriched sequences, they don't look anything like what the receptor is generally supposed to bind.”
A spark of imagination guides solution
To address this, they turned to an uncommon collaborator: Parameswaran Ramanathan, PhD, an electrical and computer engineer at the University of Wisconsin-Madison. “He said they do this sort of pattern finding all the time in his lab when they are designing electrical circuits," Ansari said. This process involves finding the shortest route with the minimum input necessary to perform a function. In the world of digital circuit design, the approach relies on defining Boolean “MinTerms”, mathematical expressions which function like road signs highlighting the different routes to a destination. “All receptor-bound DNA sequences are there, and then you prune to the absolute minimum set of MinTerms needed to achieve the task of accurately mapping genome-wide binding sites that may not contain widely recognizable DNA binding sites.”
Ansari and Ramanathan wondered if the MinTerm approach could be applied to nuclear receptor binding. The result was MinSeq Find; a search algorithm specifically designed to unravel the factors necessary for nuclear receptors to bind DNA down to the single nucleotide. “Our collaborators applied MinTerm concepts to pull out around a thousand sequences that they could use to weigh binding sites of every possible kind,” Ansari said. This allowed the researchers to investigate the specific DNA sequence combinations that determine how and why nuclear receptors bind genomes where they do.
Another bend in the tale
What they found was a finely tuned process driven not just by sequence, but also by initial ligand binding restrictions, like whether one or two receptors are needed, the importance of communication between the ligand binding domain and the DNA binding domain, and even the structure of the DNA itself. “Structure does matter,” Ansari said. “It can help distinguish between proteins that appear to bind the same DNA sequence in principle.”
These types of nuances are all born out of the DNA sequence, Ansari explains. “If the binding site has certain sequences beside it, the overall DNA binding site can be slightly bent. While some nuclear receptors couldn't care less that part of the genomic binding sites is bent, there are others that are exquisitely sensitive. In other words, if a bend occurs in the DNA, some nuclear receptors will no longer bind to the otherwise perfect site, or will bind very poorly.”
Ansari hopes that this work can help transform how we look at, and target, nuclear receptors. So much weighs on the sequences that these proteins bind to, so by identifying the non-traditional binding site properties that influence their receptor’s function, we can more successfully control this event therapeutically. This is especially true for diseases caused by aberrations in our genome. Single nucleotide polymorphisms (SNPs, pronounced “snips”) are tiny changes in our genetic code, like spelling errors. “These SNPs can kill a receptor binding site while potentially creating a binding site for something else to cause a problem,” Ansari explained.
Invaluable insight into disease pathology
Ansari has now turned his attention to what the researchers termed “orphan SNPs.” “We know of their existence, but we don't know what they do or why they might be linked to the disease,” he said. “If the SNP creates a new binding site, we can now test whether it is the receptor binding to that site that's leading to dysregulation of a disease-linked gene network or something else,” Ansari stated. “Such insights will open a new path to guide precision medicine. There are so many possibilities.” But with tools like MinSeq Find, these questions are one step closer to being answered.